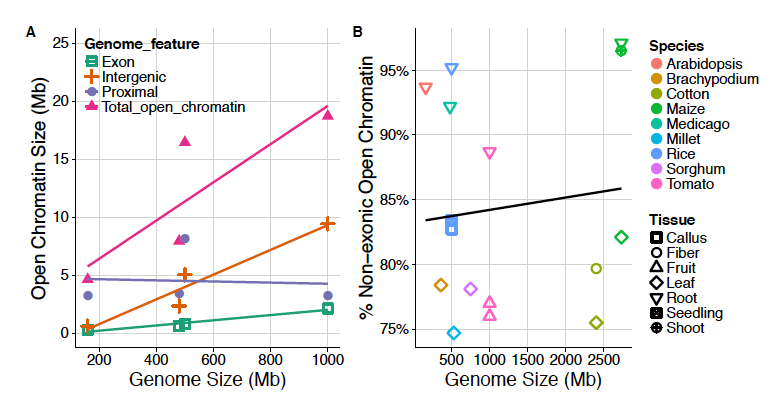
Here we propose the functional space hypothesis, positing that mutational target size scales with genome size, impacting the number, source, and genomic location of beneficial mutations that contribute to adaptation. Preliminary evidence, mostly from Arabidopsis and maize, appears to bear out at least some of the predictions of our hypothesis, but clearly more data are needed before any rigorous assessment can be made. If correct, the functional space hypothesis suggests that we should expect plants with large genomes to exhibit more functional mutations outside of genes, more regulatory variation, and likely less signal of strong selective sweeps reducing diversity. These differences have implications for how we study the evolution and development of plant genomes, from where we should look for signals of adaptation to what patterns we expect adaptation to leave in genetic diversity or gene expression data. While flowering plant genomes vary across more than three orders of magnitude in size, most studies of both functional and evolutionary genomics have focused on species in the extreme small scale. Our hypothesis predicts that methods and results from these small genomes may not replicate well as we begin explore large plant genomes.